March 29, 2021
Using deep learning for sleep stage classification
by Daniel Polimac, Data Scientist
Sleep is a crucial biological process and has long been recognized as an essential determinant of human health and performance. Whilst not all of sleep’s functions are fully understood, it is known to restore energy, promote healing, interact with the immune system, and impact both brain function and behavior. Chronic changes in sleep have been associated with a plethora of serious medical problems from obesity and diabetes to neuropsychiatric disorders. In recent years, there has been a significant expansion in the development and use of multi-modal sensors and technologies to monitor physical activity, sleep, and circadian rhythms. Deep learning approaches have gained traction in recent years for the task of classifying sleep–wake cycles and sleep stages in multi-modal sensor data. With the availability of raw PSG signals, several deep-learning techniques such as convolutional neural networks and recurrent neural networks have been used to exploit the temporal nature of this unstructured data to distinguish the sleep–wake cycles and to better understand sleep-related disorders. Today, we’ll cover how can raw PSG signals be used to classify sleep stages with deep convolutional networks. But, before, that, let’s take a quick dive into what sleep stages are and learn more about our data.

There are two basic types of sleep: rapid eye movement (REM) sleep and non-REM sleep (which has three different stages). Each is linked to specific brain waves and neuronal activity. You cycle through all stages of non-REM and REM sleep several times during a typical night, with increasingly longer, deeper REM periods occurring toward morning.
Stage 1 non-REM sleep is the changeover from wakefulness to sleep. During this short period (lasting several minutes) of relatively light sleep, your heartbeat, breathing, and eye movements slow, and your muscles relax with occasional twitches. Your brain waves begin to slow from their daytime wakefulness patterns.
Stage 2 non-REM sleep is a period of light sleep before you enter deeper sleep. Your heartbeat and breathing slow, and muscles relax even further. Your body temperature drops and your eye movements stop. Brain wave activity slows but is marked by brief bursts of electrical activity. You spend more of your repeated sleep cycles in stage 2 sleep than in other sleep stages.
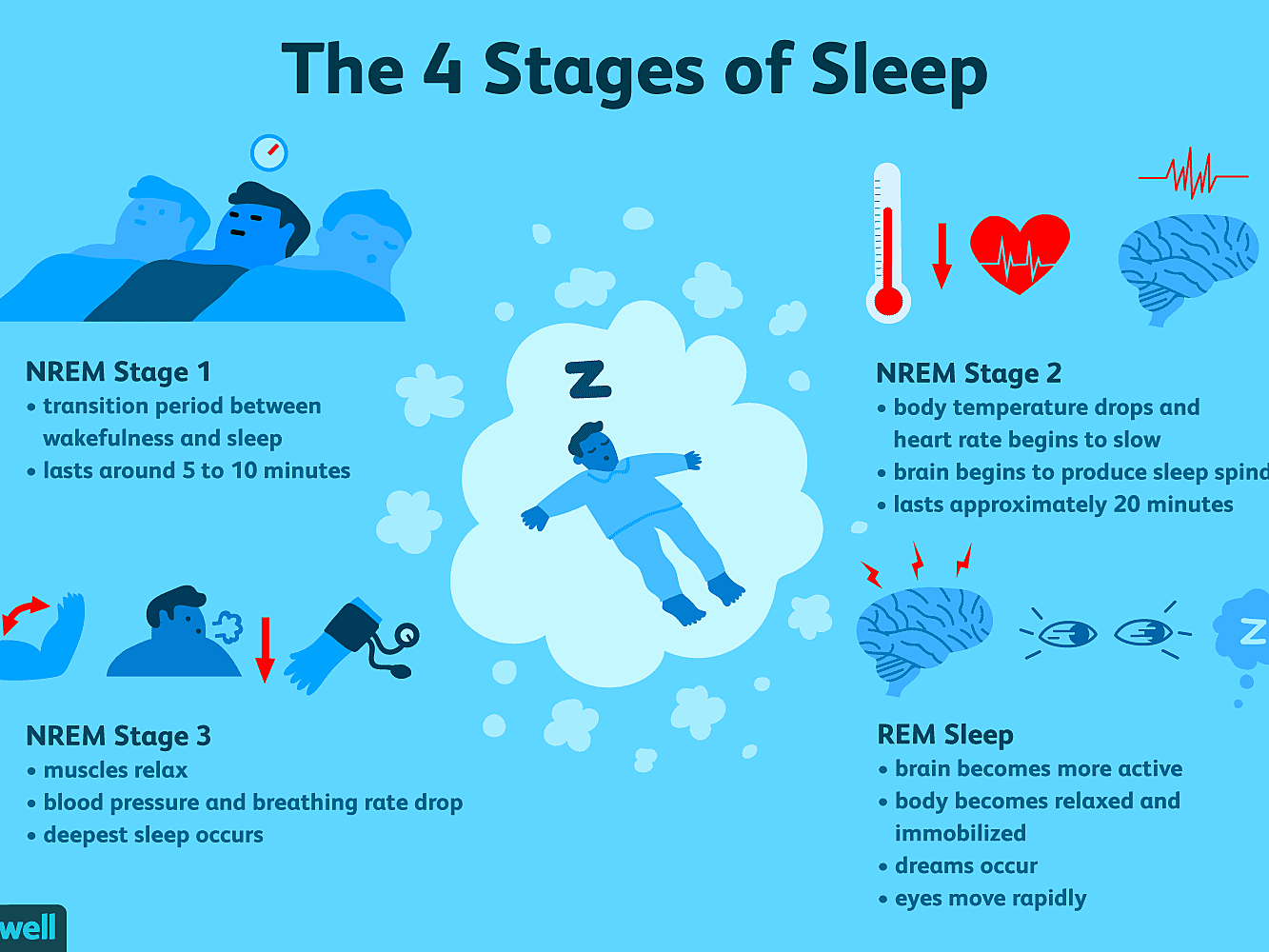
Stage 3 (often grouped with stage 4) non-REM sleep is the period of deep sleep that you need to feel refreshed in the morning. It occurs in longer periods during the first half of the night. Your heartbeat and breathing slow to their lowest levels during sleep. Your muscles are relaxed and it may be difficult to awaken you. Brain waves become even slower.
REM sleep first occurs about 90 minutes after falling asleep. Your eyes move rapidly from side to side behind closed eyelids. Mixed frequency brain wave activity becomes closer to that seen in wakefulness. Your breathing becomes faster and irregular, and your heart rate and blood pressure increase to near waking levels. Most of your dreaming occurs during REM sleep, although some can also occur in non-REM sleep. Your arm and leg muscles become temporarily paralyzed, which prevents you from acting out your dreams. As you age, you sleep less of your time in REM sleep. Memory consolidation most likely requires both non-REM and REM sleep. Throughout the night the brain goes through several 90-minute cycles of REM and Non-REM sleep. Non-REM sleep involves the sequential replay of acquired memories. In contrast, REM sleep involves a more random associate game involving disparate memories.
And what is this PSG thing?
Traditionally, polysomnography (PSG), paired with clinical evaluation, has been the gold-standard and de-facto technique to study sleep in clinical and laboratory settings, as well as to diagnose a subset of sleep disorders. PSG usually measures brain activity through electroencephalogram (EEG), airflow, breathing effort and rate, blood oxygen levels, body position, eye movement, the electrical activity of muscles, and heart rate. Traditionally, PSG requires participants to sleep in a laboratory setting. The results are then scored by an expert who has received training on how to interpret these signals. Manual sleep scoring suffers from several drawbacks. It is time-consuming, subject to biases, inconsistent, expensive and must be done offline. To counter that, machine learning and deep learning approaches have been adopted for the task of automatically classifying sleep stages from PSG data.
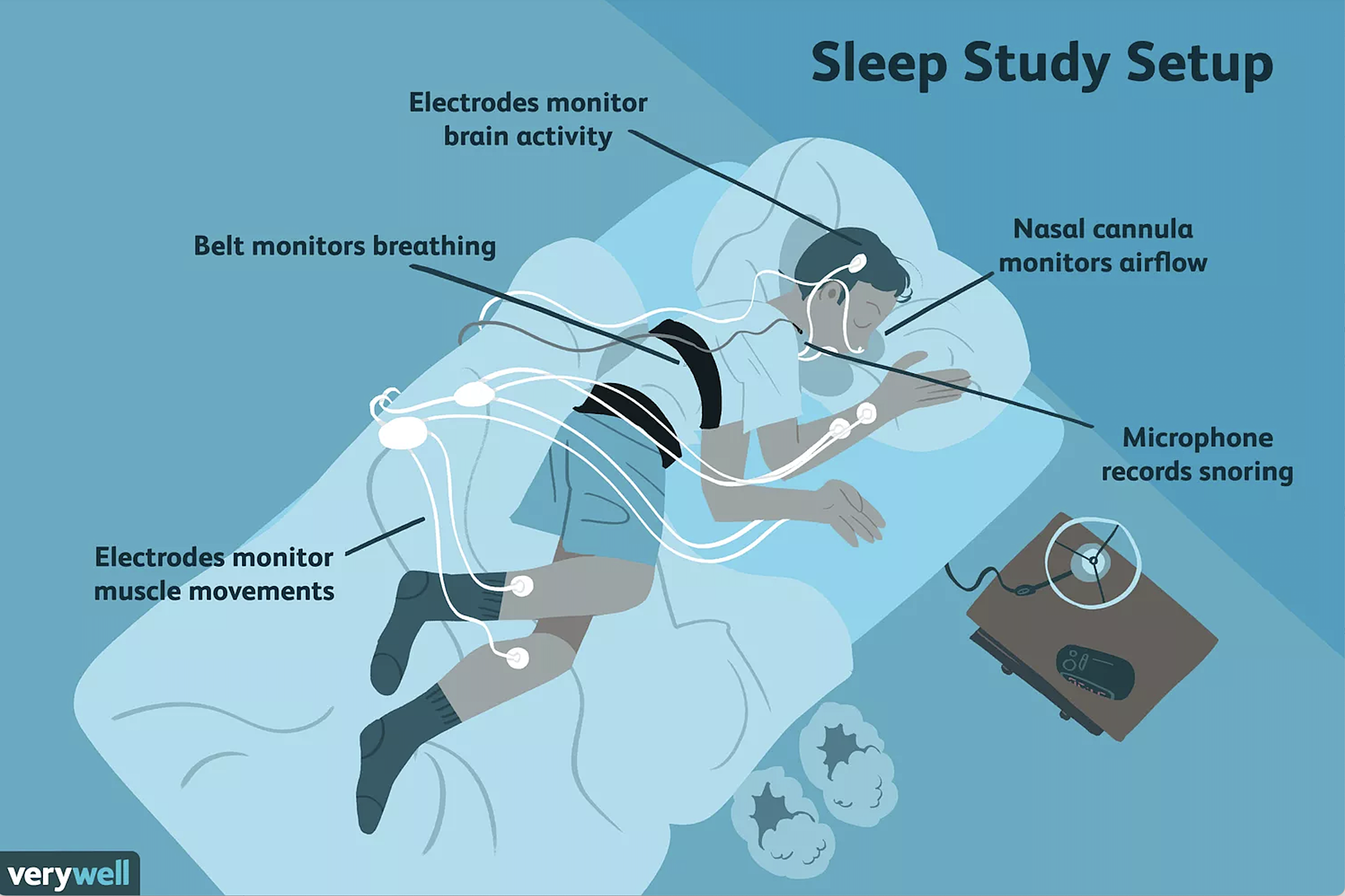
The data that we ill fetch is from the publicly available subjects from PhysioNet’s study of age effects on sleep in healthy subjects. This corresponds to a subset of 153 recordings from 37 males and 41 females that were 25-101 years old at the time of the recordings. The Physionet dataset1 is annotated using 8 labels: Wake (W), Stage 1, Stage 2, Stage 3, Stage 4 corresponding to the range from light sleep to deep sleep, REM sleep (R) where REM is the abbreviation for Rapid Eye Movement sleep, movement (M), and Stage (?) for any none scored segment. We will work only with 5 stages: Wake (W), Stage 1, Stage 2, Stage 3/4, and REM sleep (R). To do so, we use the event_id parameter to select which events are we interested in and we associate an event identifier to each of them. Moreover, the recordings contain long awake (W) regions before and after each night. To limit the impact of class imbalance, we trim each recording by only keeping 30 minutes of wake time before the first occurrence and 30 minutes after the last occurrence of sleep stages.
Before starting out, we need to import the necessary libraries.

After importing the libraries we’ll download data for 30 sleepers from the Physionet dataset, using only recordings for the first out of two nights.

After downloading the dataset, we can inspect the data for one of our sleepers. Here we can see all the channels and their values in this SG recording.
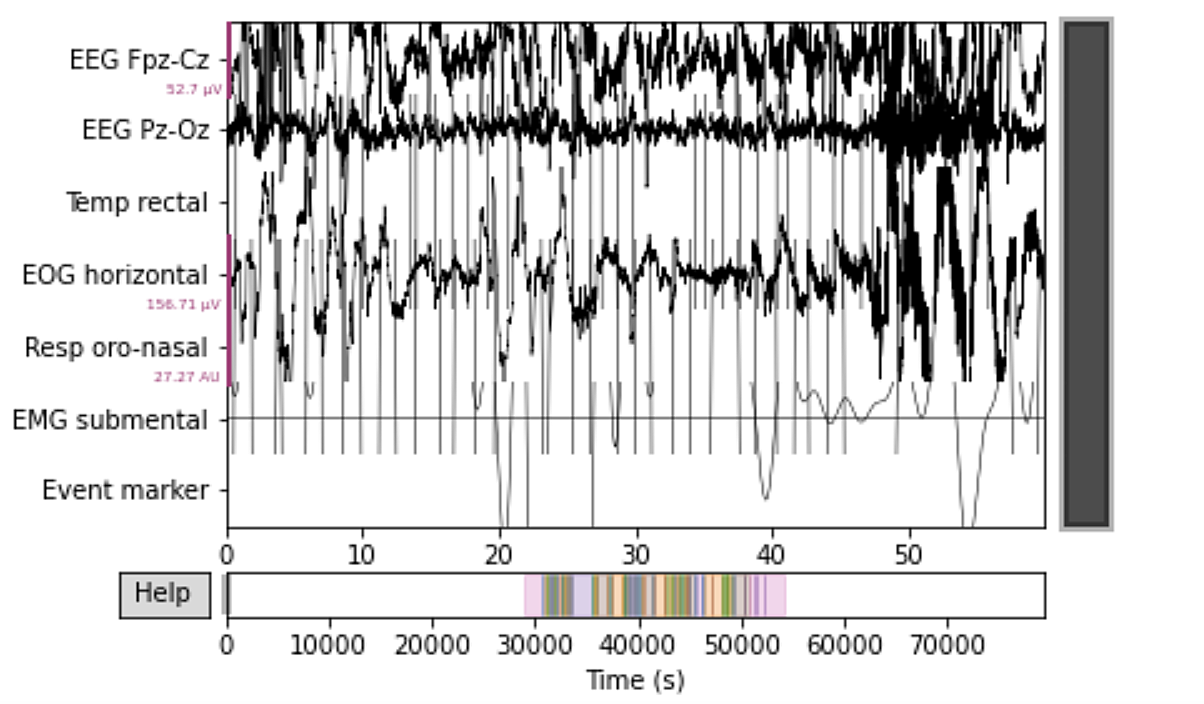
Downloaded data is in a specific format called EDF, which is an abbreviation for the European Data Format most commonly associated with PSG data files. It consists of two files of which the first contains the signal data, and the second one contains signal annotation. So the first step here is to join these two together for all 30 of our sleepers. The next step is to remove the wake stage data and leave only the first 30 minutes before and after sleep onset. After that, we create channel mappings and events id so we could reshape our data using the annotated events and the channels associated with them. An epoch is usually a timespan around an event we’re interested in. These epochs are usually 30 seconds in length and are being used by sleep technicians to evaluate sleep stages. Here, we’ll also specify our epochs to be 30 seconds. Finally, we’ll extract the data in a form of Numpy arrays and concatenate it to form a training and validation dataset. In order not to do this for each of the 30 sleepers individually we’ll create a function called signal_etl.

Let’s now run our signal_etl function and create training and validation datasets to be used with the model’s training.

Creating the model
After the data preprocessing step has been completed we can now approach creating a deep learning model for our data. We’ll model the data using 1D convolutional network architecture, generally used in sequence and time series classification. The convolutional neural network, or CNN for short, is a specialized type of neural network model designed for working with two-dimensional image data, although they can be used with three-dimensional and one-dimensional data, as with our case today. Central to the convolutional neural network is the convolutional layer that gives the network its name. This layer performs an operation called convolution. Convolution is the simple application of a filter to an input that results in activation. Repeated application of the same filter to an input results in a map of activations called a feature map, indicating the locations and strength of a detected feature in input, such as an image, a signal, or a video.
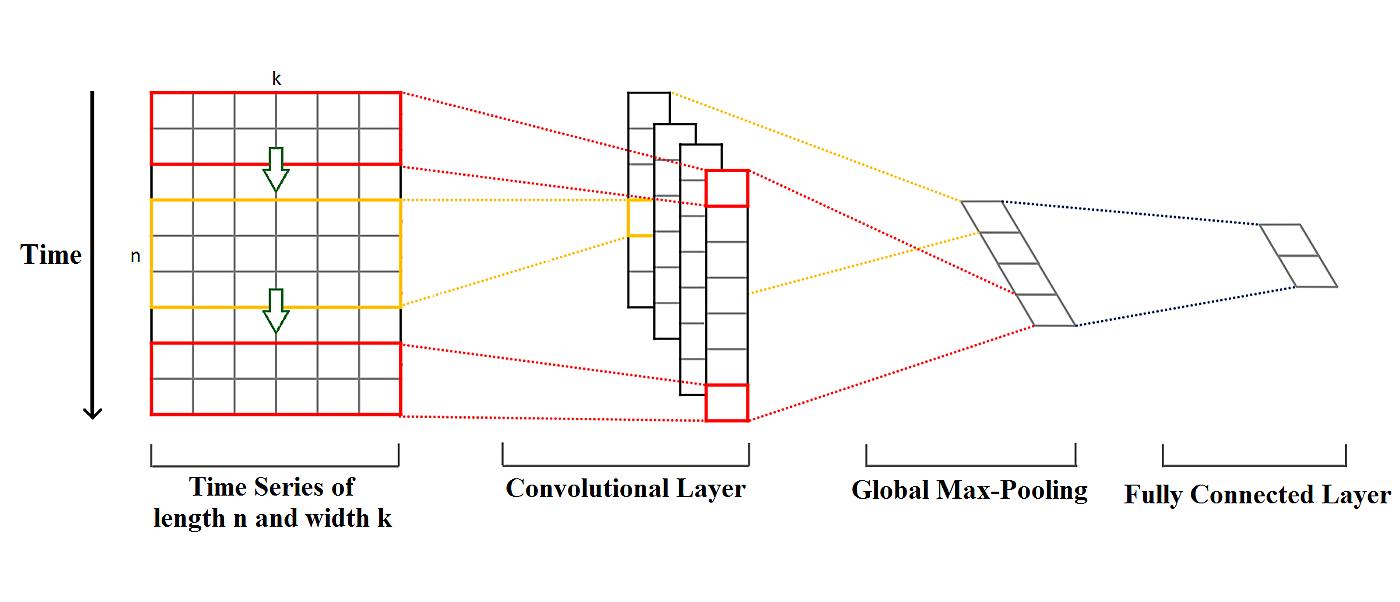
In our model, we are using three 1D convolution layers, two Batch Normalization layers, and three Max-Pooling layers. Two Dropout layers were added in between the convolutional blocks to prevent overfitting. We are also using a flatten layer to flatten our data which is then followed by a single Dense layer used for class output.
The code below is for creating our convolutional model.

Next, we need to compile the model. Here we’re going to define the loss function of the model which is going to be sparse Categorical Cross Entropy, the model’s optimizer algorithm which is going to be Adam, and our metric which is going to be Accuracy, given that we’re dealing with classification problem.
After compiling, our model is ready for training. We’ll enter the previously created x_train and y_train as our training datasets, and x_test and y_train as our validation datasets. The batch size determines the number of training examples utilized in one iteration of the training session. The large batch size can increase training speed at the cost of memory, but can often result in the model missing out on some of the more fine-grained dependencies. The number of epochs is the number of how many times will the model run through the whole dataset during the training session. Finally, we save our model to a history variably so we’ll be able to plot the training progress in terms of accuracy and loss for both training and validation.

Last, but the least here is the code for plotting the result of our training session. For the more advanced approach to monitoring the training session, I’d recommend using the Tensorboard tool.

After completing the training we are ready to see how our model is performing on our test data, i.e. the data the model hasn’t seen during training. This will tell us how accurately we can are able to classify unseen data. With this, we have successfully made a 1D Convolutional Neural Network Models for classifying sleep stages.
What happens if we’re not satisfied with our model’s performance?
Here we get into hyperparameters tuning. Hyperparameters are different parts of your model that can be tuned to affect the model globally. It’s a sort of a lock picking or radio tuning. You try different things out until you arrive to your solution. So what are some of these hyperparameters we can tune? We can change and use different activation functions in order to increase our accuracy, e.g. we can try out LeakRelu, Sigmoid, or some other activation functions and see how this affects the model’s performance. We can add more layers which will learn more complex features and will increase our accuracy. We can also vary our dropout percentage so as to avoid overfitting. Another way of avoiding overfitting is by using L2 regularization. Changing the number of epochs and also batch size will help us increase our accuracy. There are more parameters with which we can increase our accuracy but there will be a slight change in the accuracy of the model.
Concluding remarks
The impact that sleep has on human health is undeniable. Recent advances in sensing technology, big data analytics, and deep learning allow for truly ubiquitous and unobtrusive monitoring of sleep and circadian rhythms In this article you’ve learned about what sleep stages are and how deep learning can be applied to classify them. It is worth mentioning that the proposed method is not the only one that exists. There are many other approaches to both signal processing and feature generation as well as different models such as Stacked LSTMs, CNN-LSTM, ResNet, and many others. Therefore, I invite you to further explore new datasets and models, and maybe create your personalized sleep program using deep learning.
Industries
Expertise
Android
iOS
Java
Javascript
.Net
Ruby
Python
C/C++
Flutter
Angular
Blockchain
© 2025 Klika d.o.o.